Maral Maghsoudi
Machine Learning Researcher & Computational Scientist in Biomedical Data Science

I’m Maral (Zeynab) Maghsoudi, a Computer Scientist and PhD Candidate specializing in machine learning, multi-omics data integration, and pathway analysis.
With a background in software engineering and extensive experience in bioinformatics, I develop scalable, reproducible pipelines for analyzing high-dimensional omics data. My work includes co-developing the RCPA R package for consensus pathway analysis and collaborating with NASA GeneLab to investigate mitochondrial function across species in spaceflight. Currently, I focus on individual-level pathway analysis methods, applying autoencoders, matrix factorization, and other machine learning frameworks to identify personalized molecular signatures. I am also developing AI agents to support automated data integration, analysis, and knowledge discovery in complex biological systems. In parallel, I am expanding into single-cell multi-omics and exploring new approaches at the intersection of artificial intelligence, data science, and systems biology. Driven by curiosity and rigor, my goal is to bridge computer science and biology to advance precision medicine and biomedical discovery.
Education
Research Interests
Ph.D. in Computer Science
University of Nevada, Reno
GPA: 3.8/4.0
Research: Development of Computational Methods for Analyzing Single/Multi-Omics Data for Systems-Level Understanding
M.Sc. in Software Engineering
Iran University of Science and Technology
GPA: 4.0/4.0
Research: A New Hybrid Approach to Malware Detection and Classification Using Machine Learning and Behavioral Analysis
B.Sc. in Software Engineering
University of Arak, Iran
GPA: 3.4/4.0
Project: Investigating of Attacks in Computer Networks
My research focuses on the integration of multi-omics data and its applications to both bulk and single-cell datasets, with a strong emphasis on machine learning and deep learning approaches. My background is rooted in bulk multi-omics integration, where I have studied diverse integration strategies and computationally combined mRNA, methylation, and CNV profiles to derive pathway activity scores and uncover biologically meaningful insights. I am highly skilled in pathway and gene set analysis, both in developing novel methodologies, such as autoencoder-based scoring and matrix factorization, and in applying existing techniques across large-scale biological datasets to interpret molecular mechanisms at the systems level. In parallel, I am developing AI agents to support automated data integration, analysis, and interpretation, with the goal of enabling scalable and reproducible discovery workflows in complex multi-omics settings. While my primary expertise lies in the analysis of large bulk datasets, I am increasingly focused on advancing single-cell multi-omics integration through scalable, noise-robust ML/DL frameworks that address the unique challenges of single-cell data. More broadly, I am passionate about applying machine learning, statistical modeling, systems biology, and AI-driven automation to drive discoveries in precision medicine and translational research, bridging computer science and biology to enable individualized insights into health and disease.
Technical Skills
Selected Projects
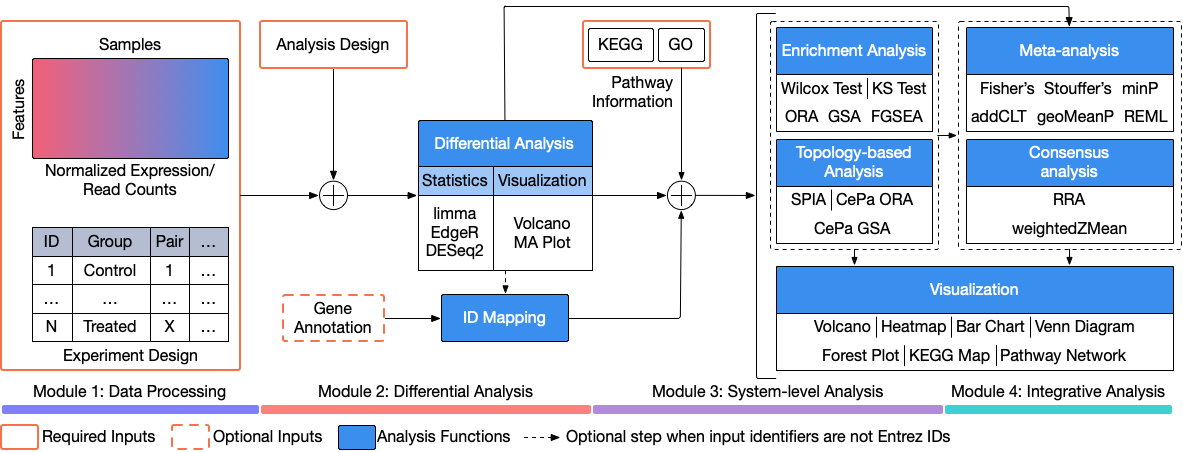
The R package for Consensus Pathway Analysis (RCPA) implements a complete analysis pipeline including: i) download and process data from NCBI Gene Expression Omnibus, ii) perform differential analysis using techniques developed for both microarray and sequencing data, iii) perform systems-level analysis using different methods for enrichment analysis and topology-based (TB) analysis, iv) perform meta-analysis and consensus analysis, and v) visualize analysis results and explore significantly impacted pathways across multiple analyses. The package supports the analysis of more than 1,000 species, two pathway databases, three differential analysis techniques, eight pathway analysis tools, six meta-analysis methods, and two consensus analysis techniques.
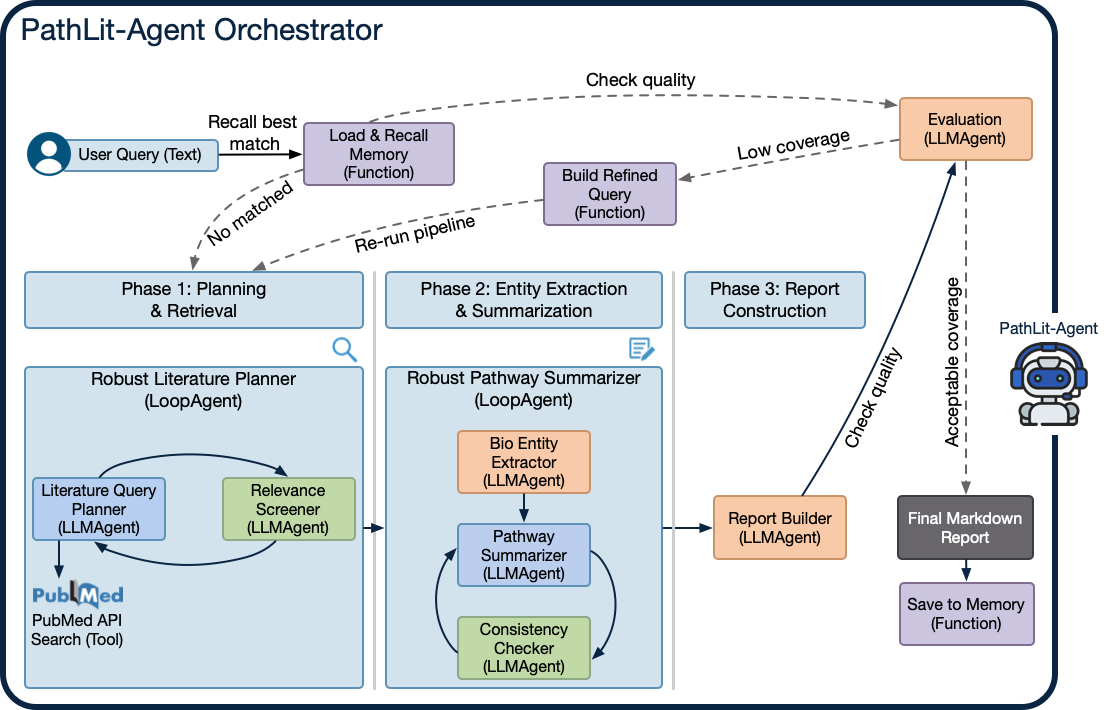
PathLit is an LLM-driven research assistant that plans literature search steps, retrieves relevant papers, summarizes findings iteratively, and generates structured biological insight. The workflow is built as a deterministic, phase-based pipeline with automated evaluation for multi-turn reliability.
Work Experience
Data Scientist & Bioinformatics Research Assistant
University of Nevada, Reno, USA
– Developed deep learning models (ResNet50, VGG-16) with advanced feature engineering (BEMD) for MRI-based breast mass classification, boosting diagnostic accuracy.
– Designed a personalized pathway analysis framework using sequential NMF for multi-omics (mRNA, methylation, CNV), improving tumor detection in TCGA data by up to 5%.
– Designed an autoencoder-based patient-level pathway analysis framework for multi-omics data, with highest accuracy for tumor detection in TCGA.
– Co-led development of the R package RCPA, enabling reproducible and scalable consensus pathway analysis workflows.
– Automated differential expression analysis for microarray/RNA-Seq with GEO support for 1,000+ species.
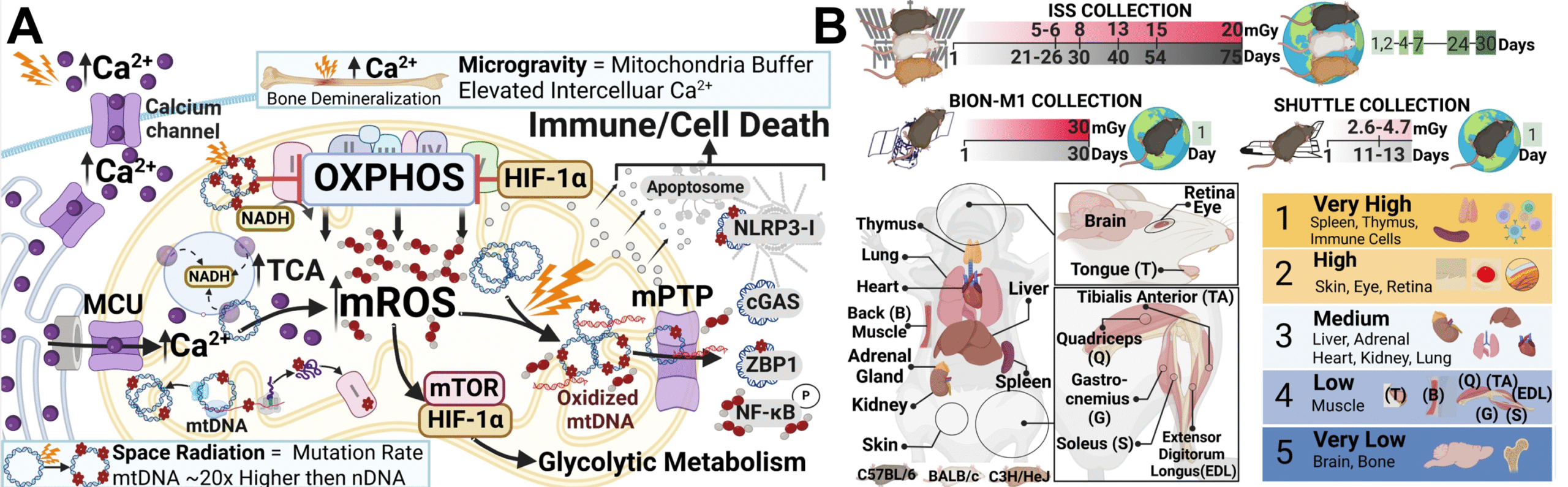
– Led pathway meta-analysis with NASA GeneLab, revealing mitochondrial dysfunction signatures across spaceflight datasets.
– Built an NGS variant calling pipeline for SARS-CoV-2 at Renown Hospital, ensuring accurate, reproducible mutation detection.
Teaching Assistant
University of Nevada, Reno, USA
– Mentored students in mitochondria-centered pathway analysis in collaboration with Purdue University Biomedical Engineering Department.
– Teaching assistant for Embedded System Design Lab for 3 years, managing and mentoring around 60 students each semester, designing lab assignments, and assisting in project-based learning.
C++ Developer | R&D Team Member
AmnPardaz, Tehran, Iran
– Researched and evaluated security solutions for antivirus tools.
– Designed, built, and maintained reliable and efficient C++ code.
– Collaborated with the software development team and provided technical feedback.
C# Developer
GoldIran (Representative of LG Products), Tehran, Iran
– Collaborated on programming the PDA to determine warehouse keeper’s tasks.
– Implemented the Warehouse Handling project to automate inventory checks.
– Collaborated on the Sales project to automate the process of taking customer purchase orders and handling further steps.
– Implemented the Soroush project to link all subsystems automatically through an automated workflow.
Malware Analysis Researcher | C# | Machine Learning
Iran University of Science and Technology, Tehran, Iran
– Developed a hybrid malware detection pipeline using static/dynamic analysis and machine learning.
– Built static analysis module to extract control flow features from system calls.
– Applied dynamic analysis using Pin and Cuckoo Sandbox for behavioral profiling.
– Enhanced malware detection performance by 7% via anti-analysis detection techniques.